The Wilson laboratory is located in the Stellar-Chance Laboratories building on the University of Pennsylvania campus in University City, Philadelphia, PA. Dr. Wilson is a faculty member in the Division of Diagnostic Innovation in the Department of Pathology and Laboratory Medicine in the Perelman School of Medicine. The Wilson laboratory uses single cell biology, spatial profiling, and computational biology as tools for investigating chronic kidney disease.
Dr. Wilson did his undergraduate training in Biomedical Engineering with a concentration in Computer Science at Johns Hopkins University. He joined the Medical Scientist Training Program at the Medical University of South Carolina where he completed his M.D. and Ph.D. in Molecular and Cellular Biology and Pathobiology. He did his residency in Anatomic and Clinical Pathology at Yale followed by a fellowship in combined Renal and Genitourinary Pathology, also at Yale. Dr. Wilson did a second fellowship in Molecular Genetic Pathology at Washington University in St. Louis before joining the faculty in the Divison of Anatomic and Molecular Pathology. His clinical expertise is in Renal and Molecular Pathology where he provides histologic and genetic diagnoses for patients with kidney disease.
Chronic kidney disease is characterized by worsening kidney function. Diabetes is the leading cause of CKD and there is a need for new therapies to prevent or slow kidney disease progression. Single cell sequencing can identify early changes in DKD that are limited to a single cell type or nephron segment. Targeting these cell-specific pathways may help to improve glycemic control and slow disease progression.
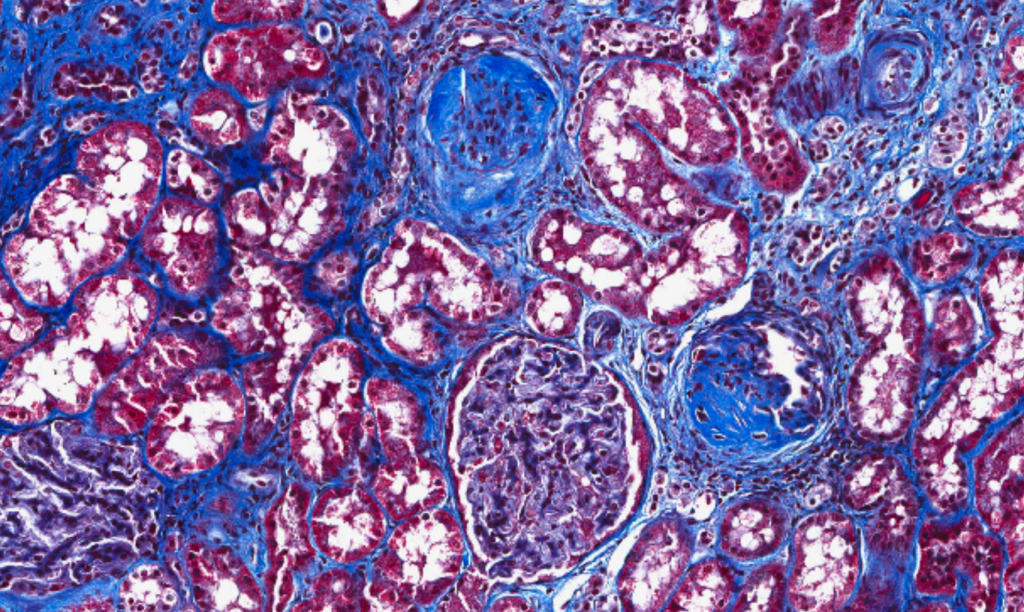 |
| Increased fibrosis in a patient with CKD. |
Single Cell Multiomics
Different cell types express different genes that determine their unique function. In the kidney, there are many cell types that work together to filter blood and produce urine in a structure called the nephron. Single cell RNA sequencing measures gene expression in every cell and this pattern can be used to determine cell type and location within the nephron. Single cell expression patterns change in acute kidney injury and chronic kidney disease and we can examine these patterns to find targetable pathways to prevent disease progression.
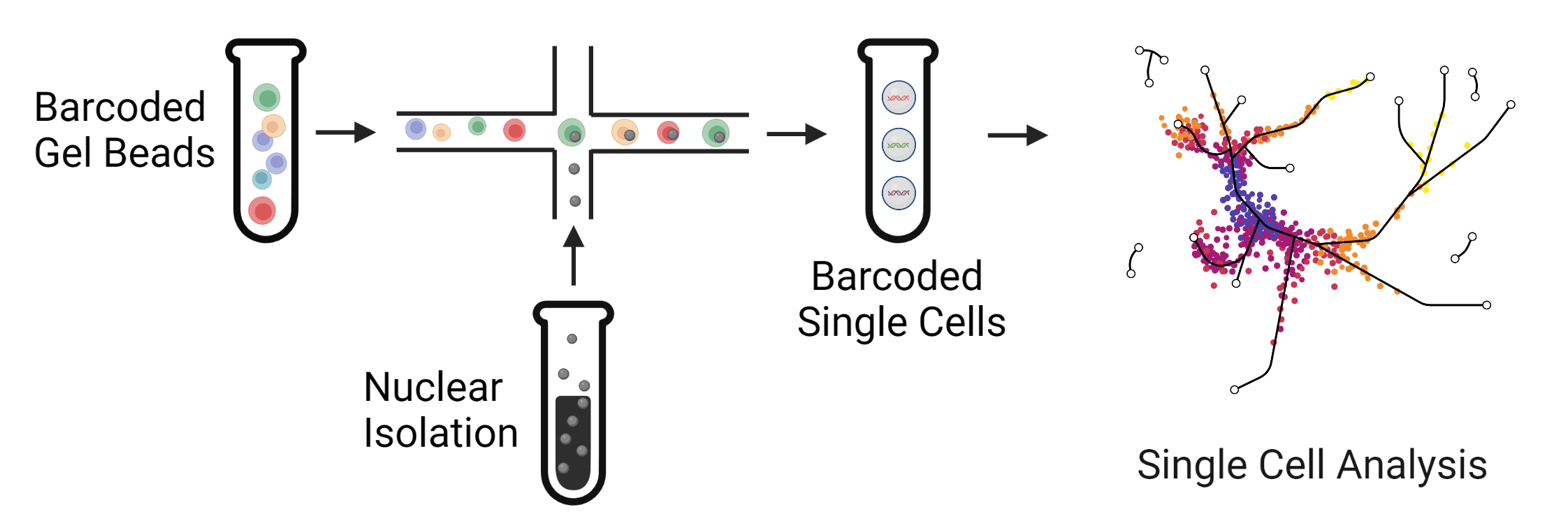 |
| Massively parallel high-throughput single cell sequencing can help to analyze thousands or even millions of cells in a single experiment. This approach helps to identify rare cell types and injured populations in kidney disease. |
Genetics in Single Cells
Genetic variation is the result of changes in the genetic code. These changes can be as small as a single base-pair or as large as an entire chromosome. We can detect genetic variants in our patient samples and ask whether these variants change expression in a specific cell type. Some of these variants may affect the function of genes that regulate kidney disease progression. The study of expression quantitative trait loci (eQTL), chromatin accessibility QTL (caQTL), and other quantitative trait loci is a rapidly-evolving field that may provide insight into why some patients with kidney disease progress more rapidly than others.
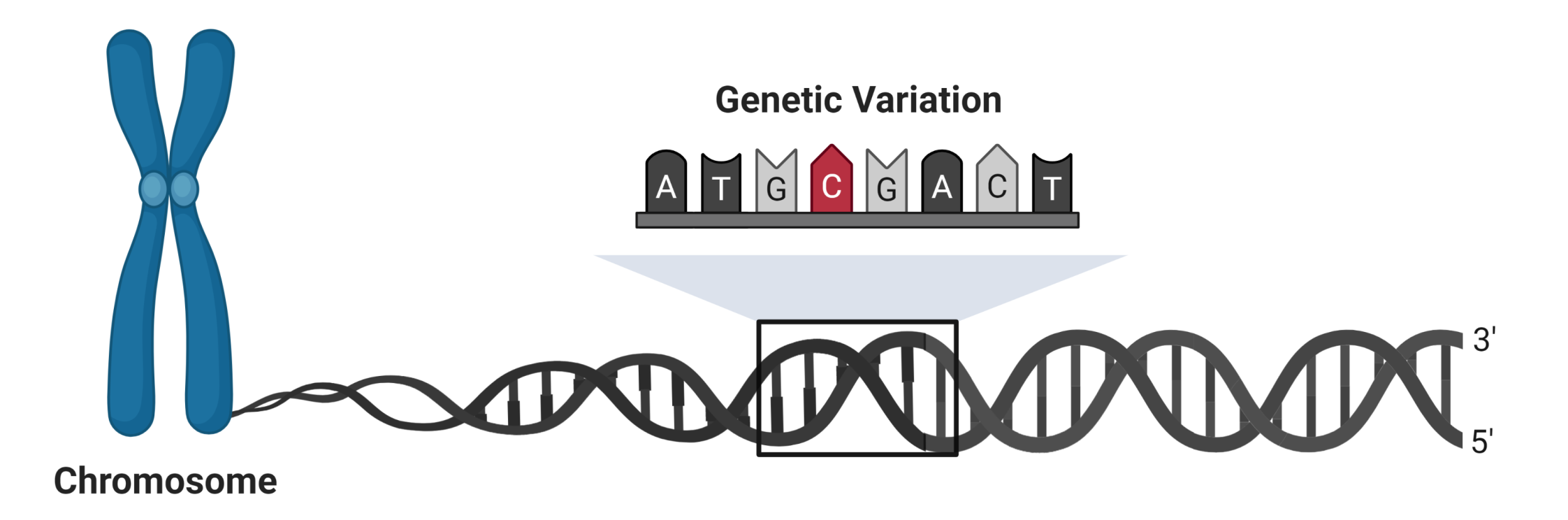 |
| Single cell sequencing can be used to examine the effect of genetic variation on kidney disease. Single nucleotide variants can modify gene expression by altering the interactions between promoters and enhancers. |
Spatial Profiling
Spatial profiling helps us to localize gene expression patterns in the kidney. Kidney disease is heterogeneous and doesn’t affect every area of the kidney in the same way. These technologies can help us to find injured cell types in the kidney to learn how tissue injury relates to kidney architecture, immune cell infiltration, and intercellular signaling that promotes kidney disease progression.
 |
| Spatial profiling can be used to localize gene expression in the kidney. The location and expression pattern of injured kidney cells can give insight into the signaling pathways that promote disease progression. |
Somatic Mosaicism
Somatic mosaicism results from the accumulation of DNA mutations over time. Some of these mutations can be as large as entire chromosome, leading to changes in hundreds or thousands of genes. We have developed digital PCR-based assays to quantify the relative ratio of chromosome copy number in complex tissues like blood and kidney. These assays can quantify the burden of somatic mosaicism and help us to understand which chromosomes are susceptible to somatic structural variation in aging and disease.
 |
| Digital PCR can be used to quantify the burden of somatic mosaicism in the kidney. Chromosomal gain and loss can give us insight into the factors that promote disease progression. |